Input parameter files¶
Instead of running wind farm optimizations via a Python script, it can also be run from input parameter
files in YAML format. For this purpose the
command line application foxes_opt_yaml has been added, following the conventions of foxes_yaml.
foxes_opt_yaml¶
Command and options¶
The command line tool foxes_opt_yaml accepts input yaml files that follow a specific
structure, that will be described shortly. A file with the name inputs.yaml can then be run in a terminal by
foxes_opt_yaml inputs.yaml
The usage is exactly the same as for foxes_yaml. The optimization specifications are simply another section that is added to the yaml file.
Input file structure¶
Let’s look at an example, available here in the foxes-opt repository:
# -----------
# foxes setup
# -----------
engine:
engine_type: process
chunk_size_states: 400
states:
states_type: ScanStates
scans:
WS: [4, 6, 8, 10]
WD: [0, 45, 90, 135, 180, 225, 270, 315]
TI: [0.05]
RHO: [1.225]
wind_farm:
layouts:
- function: add_grid
steps: [3, 3]
xy_base: [0.0, 0.0]
step_vectors: [[1300.0, 0], [0, 1300.0]]
turbine_models: [opt_yawm, yawm2yaw, NREL5MW] # Notice the opt_yawm model
algorithm:
algo_type: Downwind
wake_models: ["Bastankhah2016_linear_ka02", "CrespoHernandez_quadratic_ka04"]
wake_frame: yawed
# ------------------
# optimization setup
# ------------------
optimization:
problem:
name: opt_yawm # Name of the turbine model, wind_farm section
problem_type: OptFarmVars # Name of the optimization problem class
functions: # The following functions are called before init
- name: add_var # For this problem we need to add an opt variable
variable: YAWM
typ: float
init: 0.0
min: -30.0
max: 30.0
level: state-turbine # one variable per state and turbine
objectives: # add all objectives here to the list
- objective_type: MaxFarmPower
# constraints can be added here similarly
optimizer:
optimizer_type: Optimizer_pymoo
problem_pars:
vectorize: True
algo_pars:
type: GA
pop_size: 100
seed: 42
term_pars: [n_gen, 100]
# -------
# outputs
# -------
outputs:
# write results to netCDF file:
- output_type: SingleObjResultsWriter
functions:
- function: write_nc
fname: results.nc
# Create a rather complex plot and save it:
- output_type: plt
functions:
- function: figure
figsize: [9, 9]
result_labels: $fig
- object: $fig
functions:
- function: add_subplot
args: [2, 2, 1]
result_labels: $ax1
- function: add_subplot
args: [2, 2, 2]
polar: True
result_labels: $ax2
- function: add_subplot
args: [2, 2, 3]
polar: True
result_labels: $ax3
- function: add_subplot
args: [2, 2, 4]
polar: True
result_labels: $ax4
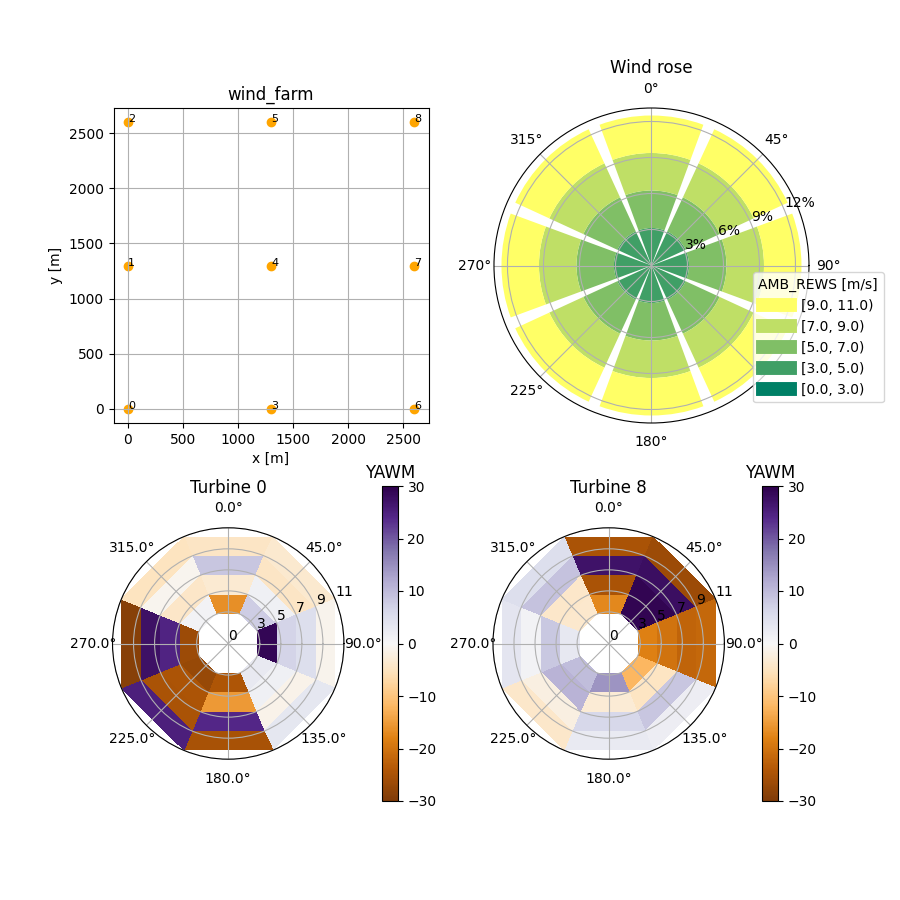
- output_type: FarmLayoutOutput
functions:
- function: get_figure
fig: $fig
ax: $ax1
- output_type: RosePlotOutput
functions:
- function: get_figure
turbine: 4
ws_var: AMB_REWS
ws_bins: [0, 3, 5, 7, 9, 11]
wd_sectors: 8
title: Wind rose
fig: $fig
ax: $ax2
- output_type: WindRoseBinPlot
functions:
- function: get_figure
turbine: 0
title: Turbine 0
variable: YAWM
ws_bins: [0, 3, 5, 7, 9, 11]
wd_sectors: 8
contraction: mean_no_weights
cmap: PuOr
vmin: -30
vmax: 30
fig: $fig
ax: $ax3
- function: get_figure
turbine: 8
title: Turbine 8
variable: YAWM
ws_bins: [0, 3, 5, 7, 9, 11]
wd_sectors: 8
contraction: mean_no_weights
cmap: PuOr
vmin: -30
vmax: 30
fig: $fig
ax: $ax4
- output_type: plt
functions:
- function: savefig
fname: result.png
- function: show
The details, especially concerning this complex output plot creation, are explained in the foxes documentation. The following image is produced by running the example: